New neural network for more accurate DNA editing
Russian bioinformaticians have proposed a new neural network architecture capable of evaluating how well a guide RNA has been chosen for a gene editing experiment. Their approach will facilitate more efficient DNA modification with the popular CRISPR/Cas method and therefore will help develop new strategies for creating genetically modified organisms and find ways of treating grave hereditary disorders. The study, supported by a Russian Science Foundation grant, was published in the Nucleic Acids Research journal.
Genomic editing, and the CRISPR/Cas method in particular, is widely used in various areas of experimental biology, as well as in agriculture and biotechnology.
CRISPR/Cas is one of the many weapons bacteria use to combat viruses. As infection occurs, the pathogen's DNA penetrates the cell, and since its sequences differ from those of the bacterium, Cas proteins recognize it as foreign hereditary material and cleave it. For the bacterium to respond to the virus faster, the cell stores fragments of the pathogen's DNA—much like a computer antivirus keeps a collection of viral signatures—and passes them on to next generations so that its Cas could thwart further attacks.
In 2011–2013, teams from different laboratories (Jennifer Doudna, Emmanuelle Charpentier, and Feng Zhang in the United States, and Virginijus Šikšnys in Lithuania) independently of one another adapted the CRISPR/Cas system to the task of introducing arbitrary changes into DNA sequences in human and animal cells, making genomic editing much easier and more efficient. The system's core elements are guide RNA, which "marks the spot," and the Cas9 protein, which cleaves DNA at that position. The cell then "mends the wound" but the changes to the genetic code have already been made.
The problem is that guide RNA targeting is not always precise and may mislead Cas9. Transforming the CRISPR/Cas technology into a practical high-precision tool is highly important, especially when it comes to medical interventions.
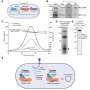
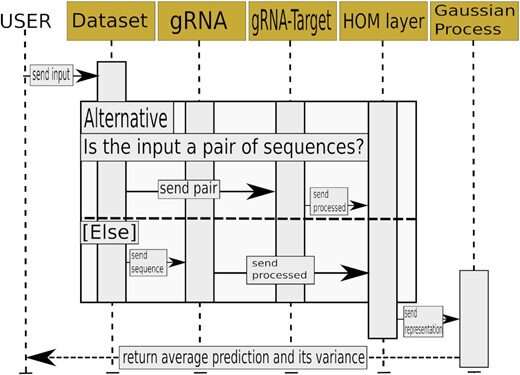
Skoltech researchers led by Konstantin Severinov have used deep learning, Gaussian processes, and other methods to make the selection of optimal guide RNAs more accurate. The team produced a set of neural networks, that is, trainable mathematical models implemented as sequential multiplication of matrices—large arrays of numbers with complex internal structure. A neural network is able to learn because it has "memory" in the form of numbers that are altered in a particular way every time the system completes a calculation in the training mode. The team trained the models on different datasets containing tens of thousands of experimentally validated guide RNAs that had displayed high-accuracy performance in human and animal cells.
The researchers proposed an algorithm that estimates the probability of DNA cleavage for a given guide RNA. The resulting scores can direct experimental design for any CRISPR/Cas-based application. The team used its neural networks to come up with a set of guide RNAs for making precise changes to the genes of the 22nd human chromosome. This has been possible thanks to the high accuracy of cleavage frequency prediction and a prediction uncertainty estimation feature, which none of the previously existing methods provided.
"Our findings can be used for a variety of CRISPR/Cas-based technology applications, such as genetic disorder treatment, farming technologies, and basic research experiments," commented Skoltech Ph.D. student Bogdan Kirillov, one of the creators of the new method and the first author of the study.
More information: Bogdan Kirillov et al, Uncertainty-aware and interpretable evaluation of Cas9–gRNA and Cas12a–gRNA specificity for fully matched and partially mismatched targets with Deep Kernel Learning, Nucleic Acids Research (2021). DOI: 10.1093/nar/gkab1065